Ginkgo.Your Innovation Partner.

Today, we’re excited to announce Ginkgo AI’s newest research release. In short, we generated a dataset measuring mRNA stability for 180,000 unique sequences, used it to train a machine learning model mapping mRNA stability to the 3’UTR sequence, then used the model to design new RNAs with improved stability.
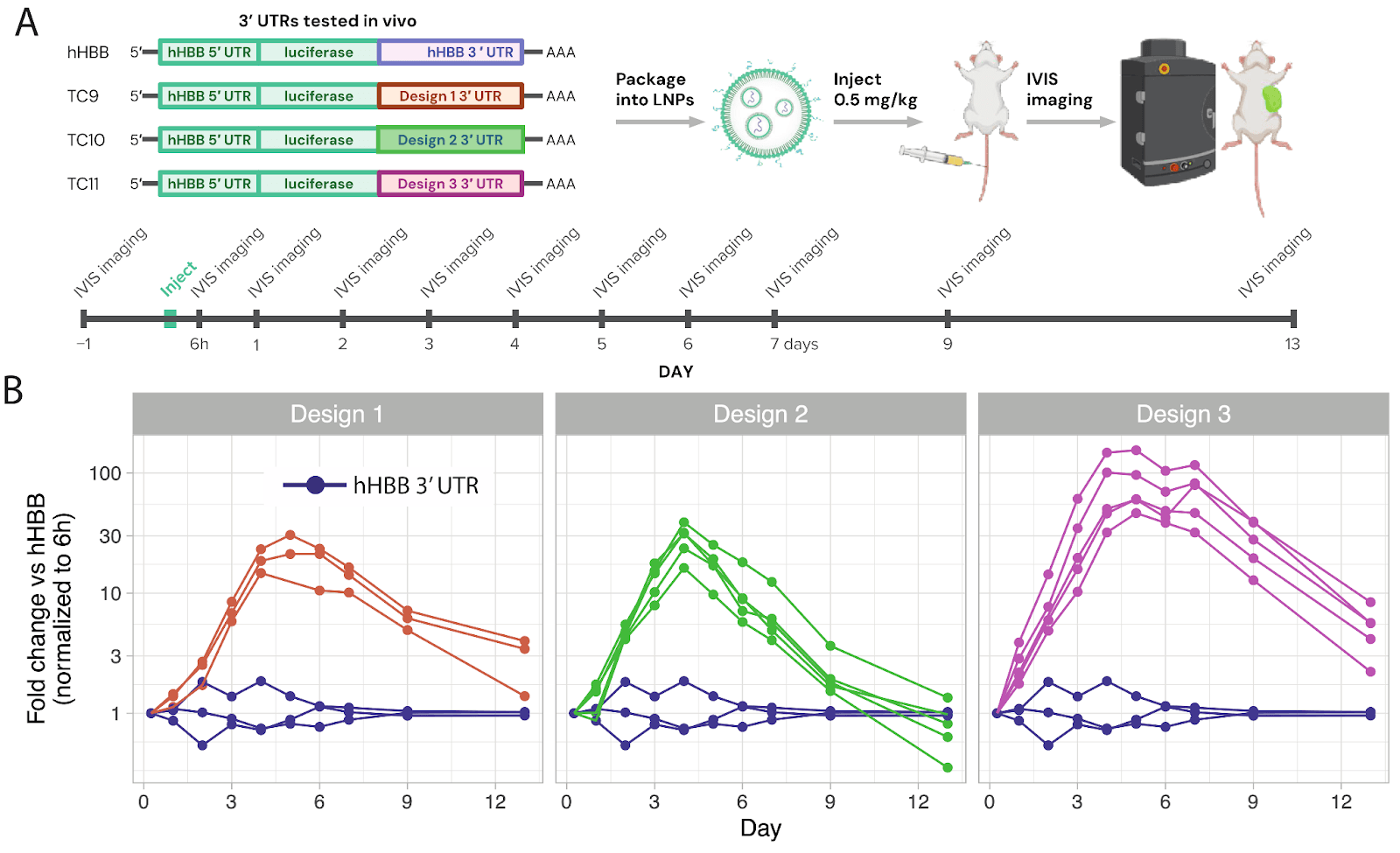
We validated designs in an in vivo mouse model, which demonstrated mRNAs that produced up to twice the protein payload over their lifetime and 30-100x more activity at the one week time point.
At Ginkgo, our mission is to make biology easier to engineer. One way we do that is by training AI models on large biological datasets. AA-0, our first protein LLM, was trained on 2 billion proprietary protein sequences and is available through our model API.
In the field of mRNA therapeutics, we saw an opportunity to use the combination of AI and large datasets to take on the long-standing problem of mRNA stability. The mRNA molecule can be notoriously fragile in vivo, limiting its ability to deliver a protein payload for therapeutics applications. In this project, we focused on designing more stable mRNA molecules by targeting the 3′ untranslated region (UTR) which are known to play a crucial role in mRNA stability.
Robust machine learning models need high-quality, comprehensive datasets for training. So, we developed a high-throughput massively parallel reporter assay (MPRA) to measure the stability of thousands of 3′ UTRs.
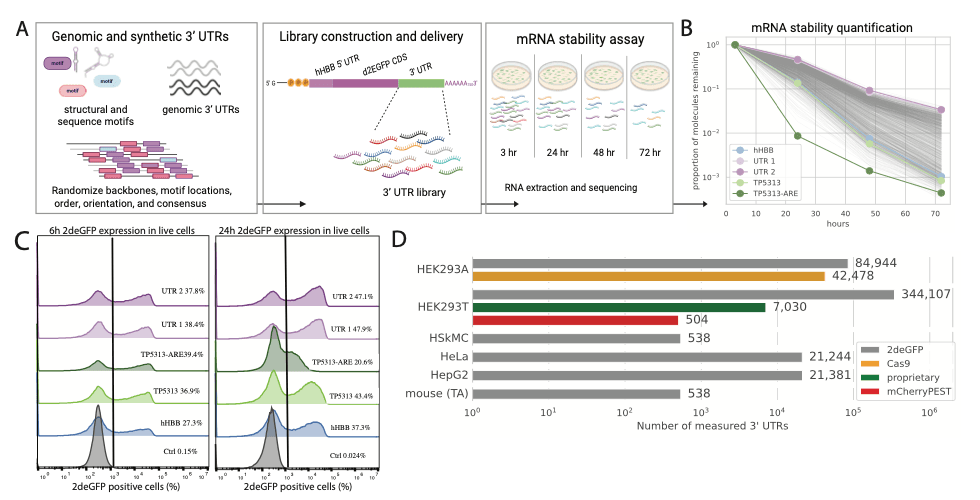
This approach allowed us to curate an extensive dataset of 180,000 unique 3’ UTR sequences and their corresponding stability measurements. Additionally, we collected a much larger dataset that measured mRNA stability of these 3’ UTRs in different cellular contexts and with different coding sequences. To our knowledge, this MPRA dataset represents the largest existing dataset relating mRNA stability to synthetic 3’UTRs – an extraordinary resource for machine learning applications.
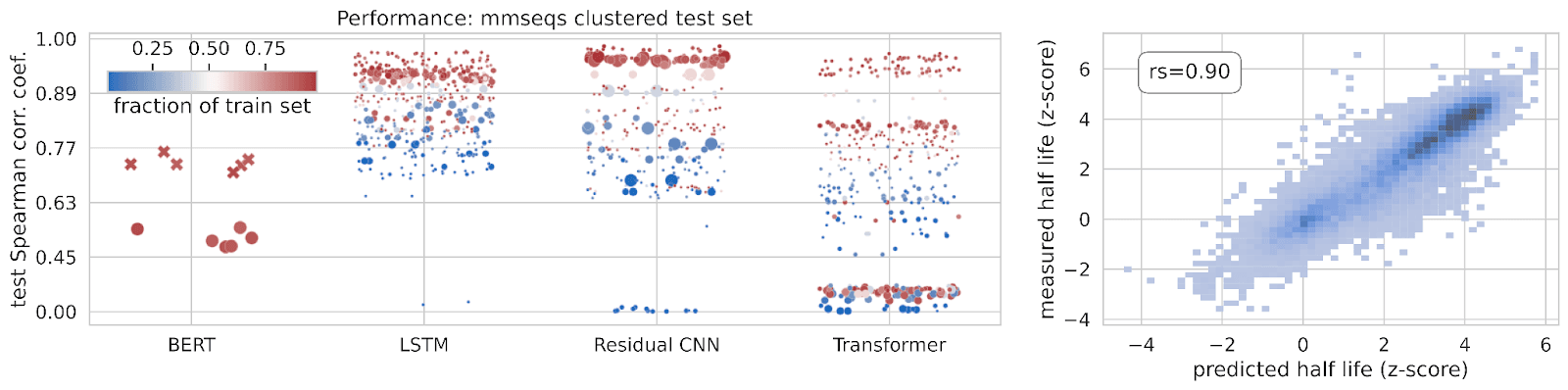
Leveraging this dataset, we trained various machine learning models to predict mRNA stability from 3′ UTR sequences and guide the design of novel synthetic 3’ UTRs with greater stability. While different types of models performed differently on their ability to predict mRNA stability from 3’ UTR sequences, we ultimately trained a long-short term (LSTM) supervised model on our MPRA data to design novel synthetic 3’ UTRs.
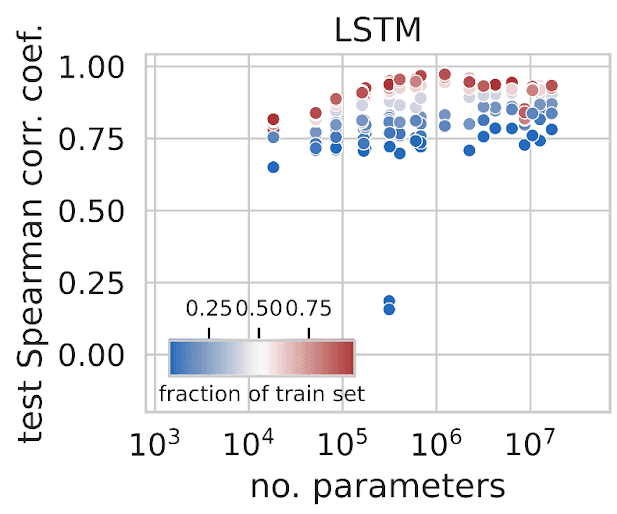
One interesting (but perhaps obvious!) finding was the effect of data quantity on model performance: the more data we trained on, the better our models performed. These findings have encouraged us to continue growing our dataset to further improve these models.
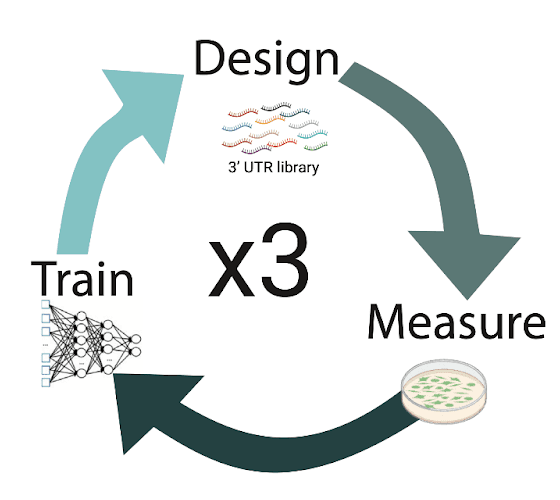
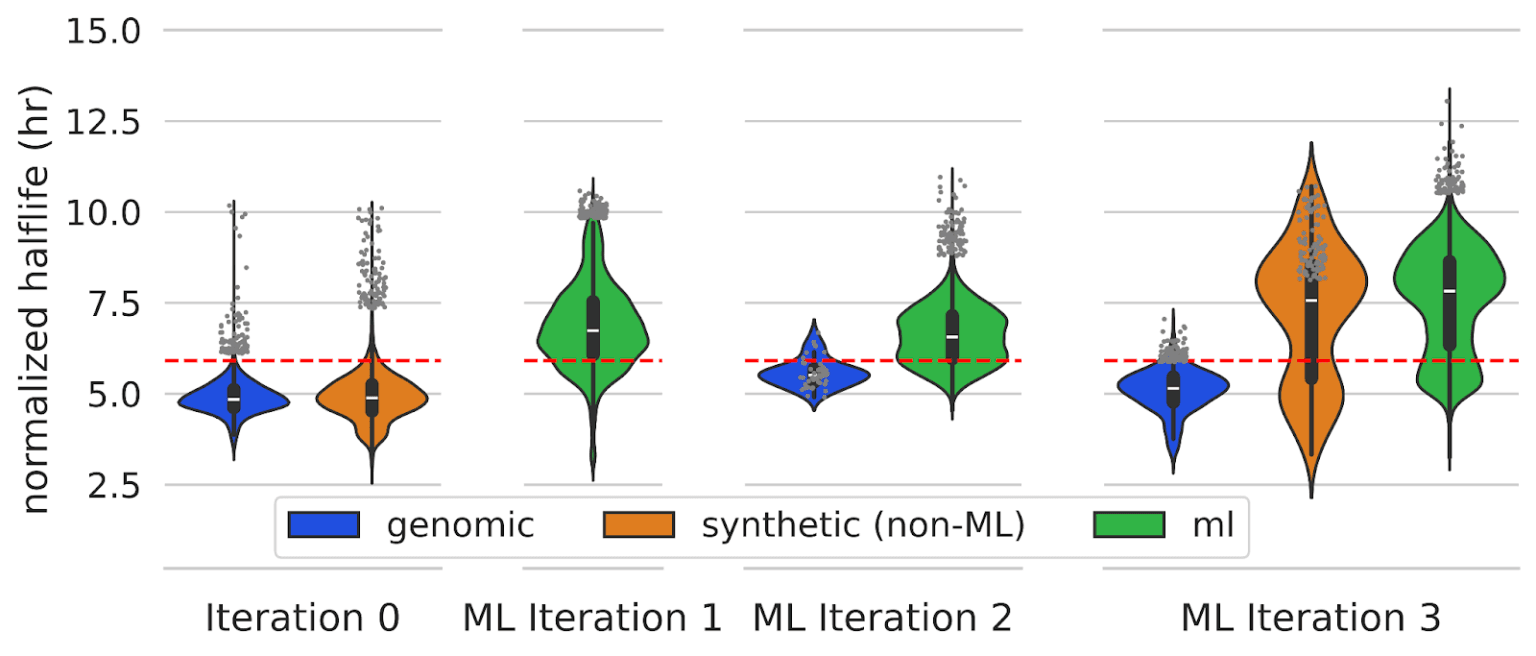
We combined our supervised model trained on stability data with various design algorithms and generative models to design novel 3’ UTRs. We ran three iterations of machine learning-driven design, and found that multiple rounds of machine learning-driven design led to significant increases in mRNA stability compared to sequences used to train our initial model. ML designs significantly outperformed a set of genomic 3’ UTRs as well as synthetic 3’ UTRs that were designed by a human.
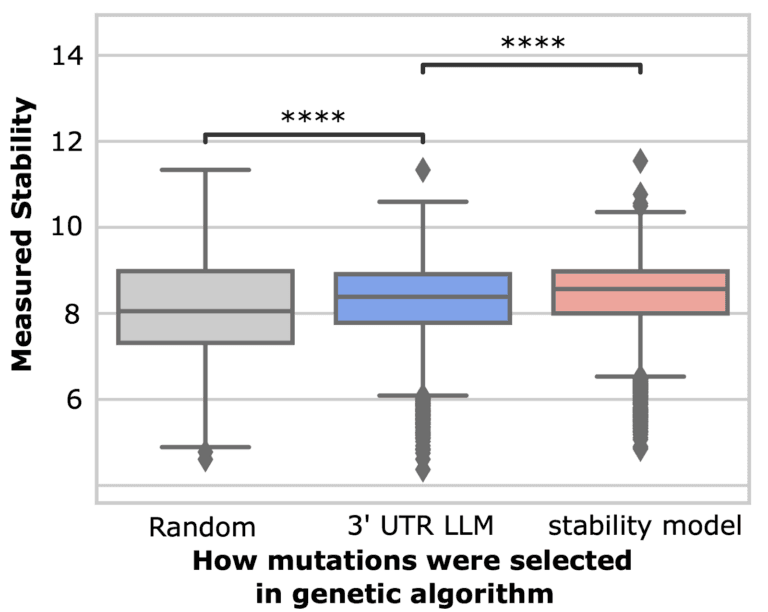
As mentioned above, we tested various strategies for designing 3’ UTRs. For one such strategy, we used a genetic algorithm to iteratively mutate and recombine 3’ UTRs, using our predictive model to score and select 3’ UTRs with greater predicted stability. We tested three methods for selecting mutations in our genetic algorithm, including random selection, selection with a 3’ UTR LLM, and selection using our supervised model. We first trained a large language model (LLM) on genomic 3’ UTRs from 125 mammalian species.
Surprisingly, we found that even though the LLM was not trained on stability measurements (only sequence), using the LLM to select the most likely mutations resulted in sequences with higher measured stability than randomly selected mutations. Our 3’ UTR LLM is available at https://models.ginkgobioworks.ai/models.
Importantly, we validated our findings by testing the ML-designed 3’ UTRs in a mouse model, resulting in up to 2-fold more protein production over time and 30-100-fold higher protein output at later time points compared to a commonly used benchmark.
The mRNA modality offers incredible versatility for therapeutic R&D. Simply by varying the sequence of the mRNA molecule, we can encode novel protein payloads with the potential to benefit human health in applications from vaccines to therapeutics.
This work shows how sequence variations in an mRNA molecule also control important aspects of its in vivo performance: stability, activity and therapeutic index. By extending the half-life of an mRNA, we might allow it to deliver more therapeutic payload at lower doses, improving efficacy and safety.
The combination of machine learning and large biological datasets offers a path towards making mRNA therapeutics easier to engineer. We’re excited about the applications this can enable, so we’re making our generative model used for 3’ UTR design trained on genomic 3’ UTRs available via our API for $0.18 per 1M tokens.
We’re entering a new era of biology. Developing datasets and models for the AI age will require a new scale of engineering. Watch this space, because we can’t wait to show you more of it.
Acknowledgements: Thanks to the members of the Ginkgo AI and Solutions teams who contributed to this work: Elise Flynn Uri Laserson JB Michel Rory Kirchner Sophia Tabchouri, Lood van Niekerk, Seth Ritter, Justin Gardin and Ankit Gupta.
Posted by Alyssa Morrow